Uncertainty in data visualisation through the lens of statistical inference
Di Cook
Monash University
Visualising Uncertainty
Rostock Retreat
June 21, 2021
https://dicook.org/files/Rostock2021/slides.html
Image credit: Di Cook, 2018
Hello 👋🏻
Hello 👋🏻

Hello 👋🏻

Hello 👋🏻

👉🏻 https://dicook.org/files/Rostock2021/slides.html
Motivation
What do you read from each display?


These images were motivated by work on the Australian Cancer Atlas, and show thyroid cancer incidence as a choropleth map (left) and a new type of display a hexagon tile map (right).
Kobakian and Cook (unpublished) https://github.com/srkobakian/experiment
What do you read from each display?
High thyroid cancer incidence mostly located in east coast.
High thyroid cancer incidence is evident around Brisbane, Sydney, Perth and in some inner city Melbourne areas.
"Land doesn’t get cancer, people do"
We need to establish a new display for Australia to allow us to read spatial distribution of values measured on people
To test which design is better we are going to use the lineup protocol. Each slide shows 12 plots, numbered 1 through 12 at the top of each plot. One of the 12 is a data plot and the remaining 11 are null plots.
To test which design is better we are going to use the lineup protocol. Each slide shows 12 plots, numbered 1 through 12 at the top of each plot. One of the 12 is a data plot and the remaining 11 are null plots.
Pick the plot that is most different from the others.
Write them down just for yourself, without sharing, for now.
To test which design is better we are going to use the lineup protocol. Each slide shows 12 plots, numbered 1 through 12 at the top of each plot. One of the 12 is a data plot and the remaining 11 are null plots.
Pick the plot that is most different from the others.
Write them down just for yourself, without sharing, for now.
Ready?
To test which design is better we are going to use the lineup protocol. Each slide shows 12 plots, numbered 1 through 12 at the top of each plot. One of the 12 is a data plot and the remaining 11 are null plots.
Pick the plot that is most different from the others.
Write them down just for yourself, without sharing, for now.
Ready?
Check your choices
| page | location |
|---|---|
| 8 | 6 |
| 9 | 3 |
| 10 | 8 |
| 11 | 5 |
| 12 | 9 |
| 14 | 3 |
Conducting visual inference
About the lineup protocol
Based on the statistical justice system
- Compare the data plot (accused) with a population of null plots (innocents)
- If the data plot (accused) is picked from the lineup, then reject the null hypothesis because it looks different (guilty).
- The p-value is the probability that the accused would look this different (guilty) if they actually were not really different (innocent).
Wickham et al (2010) IEEE TVCG
| plot | question | null |
|---|---|---|
| Chloropleth maps | Is there a spatial trend? | No relationship between location and statistic value |
| Tag cloud | Is this document the same as that document? | No difference in word counts |
| Treemap | Is the distribution within higher-level categories the same? | No difference in proportions within categories |
| Histogram | Is the underlying distribution smooth? | Distribution is smooth |
| Histogram | Is the underlying distribution bell-shaped? | Distribution is normal |
| Residual plot | Are residuals normally distributed? | Distribution is normal |
| Scatterplot | Are the two variables associated? | No association |
| Scatterplot, coloured | Are points clustered by colour? | No difference between coloured groups |
| Time series | Does the mean change over time? | Same mean over time |
| Time series | Does the variability change over time? | Same variability over time |
A visual t-test
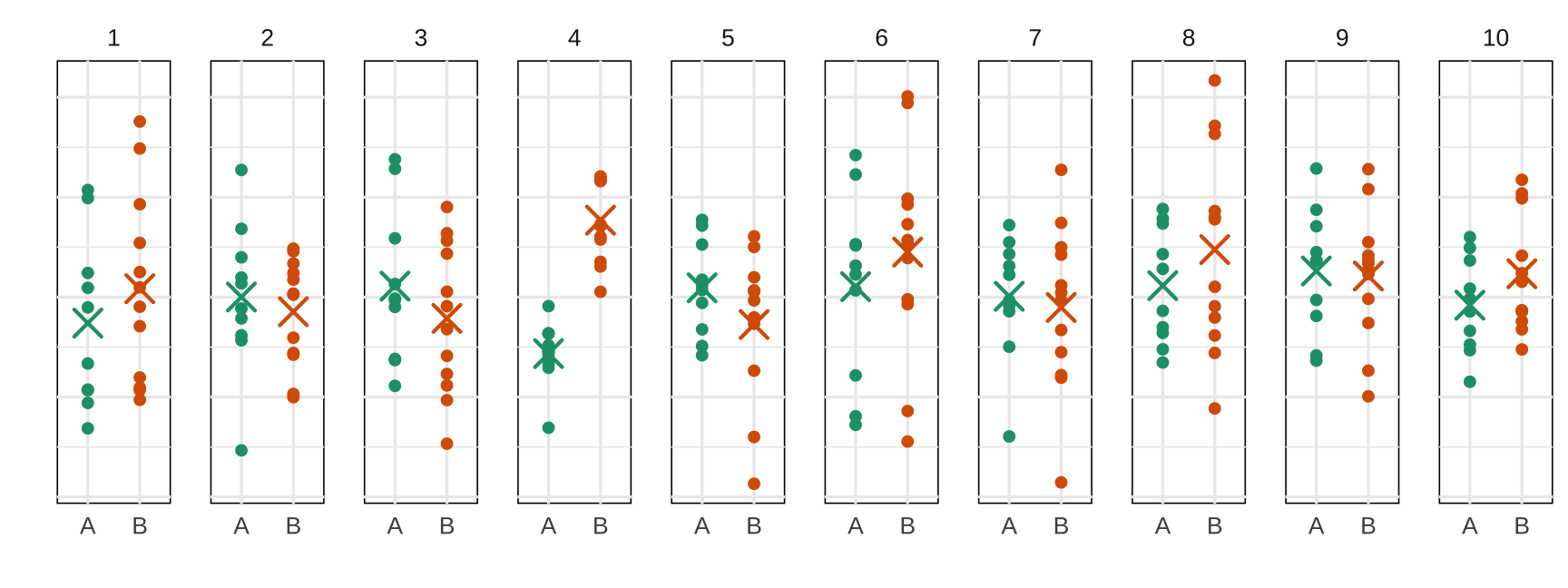
Nulls: samples simulated from same normal distribution
Null hypothesis: Both groups are samples from the same distribution
A visual t-test: take 2
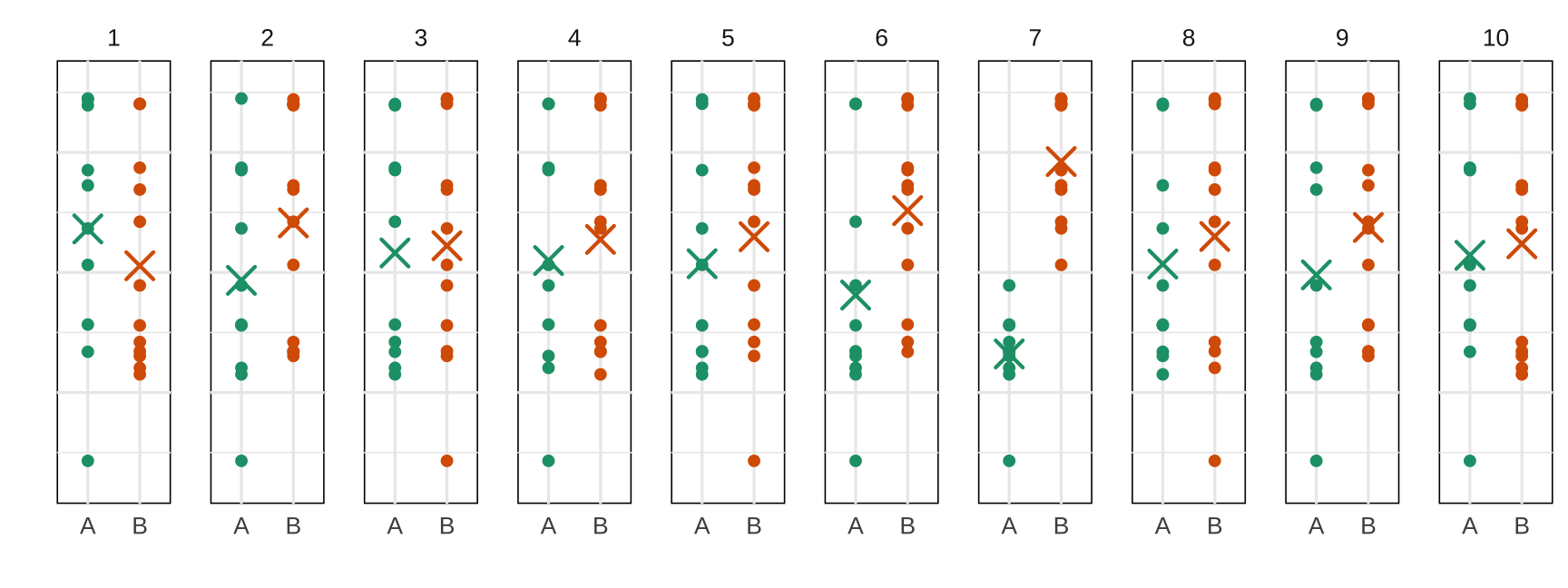
Nulls: samples generated by permuting labels A, B
Null hypothesis: Both groups are samples from the same distribution
Procedure (1/5)
| x | y |
|---|---|
| B | 0.92 |
| B | 2.18 |
| A | -0.27 |
| A | -3.92 |
| A | -2.12 |
| B | 1.73 |
| A | -1.65 |
| B | 1.81 |
| B | 3.51 |
| B | 3.60 |
| ID | koala_NSW | koala_VIC | bilby_NSW | bilby_VIC |
|---|---|---|---|---|
| grey | 23 | 43 | 11 | 8 |
| cream | 56 | 89 | 22 | 17 |
| white | 35 | 72 | 13 | 6 |
| black | 28 | 44 | 19 | 16 |
| taupe | 25 | 37 | 21 | 12 |
- Variables in columns
- Observations in rows
Procedure (2/5)
Use the grammar of graphics to define a plot
ggplot(aes(x = x, y = y, colour = x)) + geom_point() + stat_summary(fun = "mean", shape = 4, size=1) + scale_colour_brewer("", palette="Dark2") + some_nice_plot_stylingBased on this mapping, what would be considered null (not interesting)?
Procedure (3/5)
Add data or null
data %>% ggplot(aes(x = x, y = y, colour = x)) + geom_point() + stat_summary(fun = "mean", shape = 4, size=1) + scale_colour_brewer("", palette="Dark2") + some_nice_plot_stylingProcedure (4/5)
Hide the data among a field of nulls
library(nullabor)lineup(null_permute("y"), n=10, true = data) %>% ggplot(aes(x = x, y = y, colour = x)) + geom_point() + stat_summary(fun = "mean", shape = 4, size=1) + scale_colour_brewer("", palette="Dark2") + facet_wrap(~.sample) + some_nice_plot_stylingProcedure (5/5)
Ask uninvolved, independent observers to pick the plot that is most different from the lineup. We would expect that the chance that any observer chooses the data plot is 1/10 (or 1/m generally, where m is the number of plots in the lineup.
Suppose you had 23 observers, and 8 of them choose the data plot as the most different.
pvisual(x=8, K=23, m=10)## x simulated binom## [1,] 8 0.0099 0.001229827How do we know it works?
Validation experiment
Majumder et al (2013) conducted validation study to compare the performance of the lineup protocol, assessed by human evaluators, in comparison to the classical test, using subjects employed with Amazon's Mechanical Turk.
Power analysis of human evaluation relative to classical test.


How can it be used to compare plot designs?
Power to compare plot design
Hofmann et al (2012) show how power, interpreted as proportion of observers who detect the data plot, from different plot designs, can be used to establish which is better.
How to make the power calculations and generate confidence intervals for power of different designs.



Back to the maps experiment
Thyroid cancer incidence in Australia


Experimental design
Factor: choropleth, hexagon tile map
Structure: NW to SE trend, hotspot in 3 cities, hotspot in all cities
Null: Simulation from a variogram model to model spatial dependence, using gstat package (144 null sets)
Replicates: Four
Lineups: 12 plots in a lineup
Data plot: Trend added to null, for one plot in a lineup
Displays: Two sets, with coin flip for data displayed as choropleth or hexagon tile map, sets A and B
Subjects: 42 subjects for set A, and 53 for set B
Results
How to do this yourself
Get a copy of the nullabor package
install.packages("nullabor")or
# install.packages("remotes")remotes::install_github("dicook/nullabor")Look at the "Get started" documentation at http://dicook.github.io/nullabor/index.html
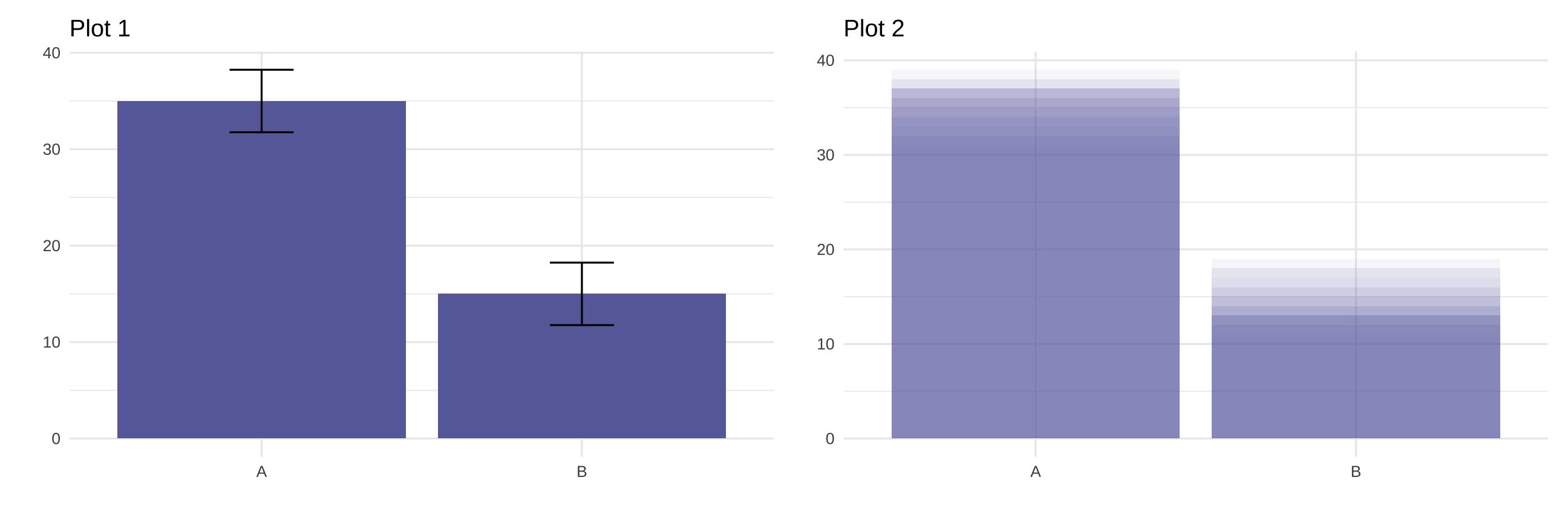
Statistical inference architecture
is built to measure uncertainty
Statistical
inference
architecture
Group A1 Data vis challenge
Which display of uncertainty is better?
Thanks for listening!
Here's what I hope you take away from this talk:
- Uncertainty means what might change if you had a different sample
- This is what statistical inference addresses
- Plots can be embedded into an inferential framework
- Crowd-sourcing can help with conducting inference with plots
- Visual inference might be helpful to test your new ideas for adding indications of uncertainty into your displays
Additional reading
^ Buja et al (2009) Statistical Inference for Exploratory Data Analysis and Model Diagnostics, RSPT A
^ Wickham et al (2010) Graphical Inference for Infovis, TVCG
^ Hofmann et al (2012) Graphical Tests for Power Comparison of Competing Design, TVCG
^ Majumder et al (2013) Validation of Visual Statistical Inference, Applied to Linear Models, JASA
^ Yin et al (2013) Visual Mining Methods for RNA-Seq data: Examining Data structure, Understanding Dispersion estimation and Significance Testing, JDMGP
^ Zhao, et al (2014) Mind Reading: Using An Eye-tracker To See How People Are Looking At Lineups, IJITA
^ Lin et al (2015) Does host-plant diversity explain species richness in insects? Ecological Entomology
^ Roy Chowdhury et al (2015) Using Visual Statistical Inference to Better Understand Random Class Separations in High Dimension, Low Sample Size Data, CS
^ Loy et al (2017) Model Choice and Diagnostics for Linear,
Mixed-Effects Models Using Statistics on Street Corners, JCGS
^ Roy Chowdhury et al (2018) Measuring Lineup Difficulty By Matching Distance Metrics with Subject Choices in Crowd- Sourced Data, JCGS
^ Vanderplas et al (2020) Testing Statistical Charts: What Makes a Good Graph? ARSIA
^ Vanderplas et al (2021) Statistical significance calculations for scenarios in visual inference. Stat.
Acknowledgements
Slides created via the R package xaringan, with wattle theme created from xaringanthemer.
The chakra comes from remark.js, knitr, and R Markdown.
Slides are available at https://dicook.org/files/Rostock2021/slides.html and supporting files at https://github.com/dicook/Rostock2021.

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Image credit: Di Cook, 2018